PTPT, the freeware program for permutation testing concordance between phylogeny and the distribution of phenetic traits.
Jaroslav Flegr1) & Pavel Záboj2)
1) Department of Parasitology, Faculty of Science, Charles University, Viničná 7, CZ-128 44 Praha 7 , Czech Republic
2} Department of Algebra, Faculty of Mathematics and Physics, Charles University, Sokolovská 83, CZ-170 00, Praha 7, Czech Republic
Permutation test, phylogeny, phylogenetic tree, program, parasite, Toxoplasma, Trichomonas
Abstract. An indispensable step of any comparative study is the testing of a concordance between distribution of phenetic traits and evolutionary history of the taxon under study. We present a computer program PTPT which can perform these testing on the basis of a permutation tail probability test. The program that can also perform the permutation tests analogical to t-test, analysis of variance (ANOVA) and to correlation analysis is available at http://www.karlin.mff.cuni.cz/~zaboj/ptpt.
The testing of concordance between the distribution of a particular trait and the evolutionary history of a taxon is a principal task of all comparative studies. The distribution of a trait within the taxon can reflect either distribution of a common function and therefore of a common selective pressure pattern (the species subjected to the same selective pressure have the same trait) or the evolutionary history of the taxon (the phylogenetically relative species share the traits) (Harvey & Pagel, 1991). The existence of statistically significant association between the distribution of the trait and the position of the taxon within the genealogical tree indicates the validity of a null hypothesis, i.e., the distribution of the trait simply reflects a random process - the evolutionary history of the taxon cladogenesis (Archie 1989).
Several approaches are being used for testing this null hypothesis depending on the type of data (character set/distance matrices) of the studied trait. If the trait is described by character data, a cladistic analysis can be performed with forced tree topology (reflecting the already known cladogenesis of the taxon). The consistency index provided by common cladistic programs can be used as simple measure of the degree of concordance between distribution of the trait and the phylogeny. The null model can be tested by a permutation tail probability test (Moore et al., 1994). If the trait is described by distance data, Mantel tests can be used to test one or more hypotheses (independent variables represented as matrices) against an observed pattern (dependent matrix) using (partial) regression or correlation (Thorpe, 1996).
The later method is more universal because any character data can be transformed to distance matrices. However, before analysis the distances should be corrected for differences in rates of evolution in different branches of phylogram. Moreover, no integrated software is available neither for Mantel test nor for other important types permutation tests.
Recently we developed program PTPT for various types of permutation tail probability tests, including those for analysis of concordance between distribution of traits and phylogeny. The program can analyze the qualitative and quantitative character data as well as the distance matrices. The phylogenetic tree can be entered in usual parenthetical format. The average distance between sister OTUs (operational taxonomic units, i.e., sister strains or sister branches of the tree) is calculated (or read from a distance matrix) and used as a measure of concordance which is tested in one-sided or two-sided permutation tail test (Manly 1991). The program can either generate all possible permutations of terminal branches of the tree or the number of trees to be generated can be user-defined. Usually 5000 random trees provide stable estimation of p-value and can be generated by ordinary PC within seconds.
EXAMPLE 1
Concordance between the pathogenicity and genealogical relationship of Trichomonas vaginalis strains
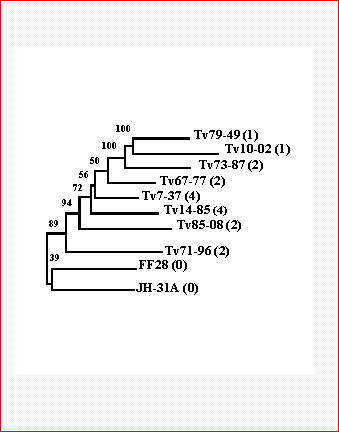
Pathogenic effects of ten T. vaginalis strains on donor female patients were assessed by clinical and histopathological findings and rated by five arbitrary units (0 - no effect, 4 - the most severe effects) (Kulda, 1989) (page 148). The phylogenetic (genealogical) tree of trichomonad strains obtained from DNA-fingerprinting data by Neighbor Joining method was ((((((((Tv79-49 Tv10-02) Tv73-87) Tv67-77) Tv7-37) Tv14-85) Tv 85-08) Tv71-96) (FF28 JH-31A)) and the pathogenicity indexes were 1, 1, 2, 2, 4, 4, 2, 2, 0 and 0, respectively (Fig. 1). The concordance between position of the strain within the tree and the pathogenicity indexes was estimated by a permutation test.
At first we can run the PTPT program with the option -h to receive a help screen:
input:
>ptpt -h
output:
usage PTPT [option] [file]
-h help
-v verbose, print partial results
-V Verbose, print permutations too
-s suppress output of final results
-m seed set seed for random generator, >=0
-n nr set number of random permutations, >=0
-a generate and test all permutations, overrides -n
-o file redirect output to file
-x variance test, -M is ignored
-c correlation test, -M is ignored
-M file read matrix file
With no FILE read standard input
Then we can test the concordance using standard (prompt line) input:
>ptpt -n 5000 <ENTER>
the program waits for standard input, we can type a tree with pathogenicity indexes: ((((((((1 1)2)2)4)4)2)2)(0 0)) <ENTER>
Within seconds we will obtain the results:
Results: 10.2738
Less: 101 2.02%
Equal: 1 0.02%
Greater 4898 98%
Alternatively, we can prepare the text file PATHO containing the tree with pathogenicity indexes and then run the test typing the following command:
>ptpt -n 20000 PATHO <ENTER>
We will obtain practically identical results, 426 (2.13%) less, 3 (0.015%) equal and 19571 (97.9%) greater. The results indicate that there is only approximately 2% probability of obtaining the same or better concordance between pathogenicity of T. vaginalis strain and its position within the genealogical tree by chance. Therefore, our experimental data suggest that phylogeneticaly related strains of T. vaginalalis express similar pathogenic effects.
Fig. 1
Phylogenetic tree for ten strains of Trichomonas vaginalis. The numbers in parentheses indicate the pathogenicity indexes (Kulda, 1989) (page 148).
EXAMPLE 2
Influence of Toxoplasma gondii infection on human personality
The program PTPT can also perform the permutation tests analogical to t-test, analysis of variance (ANOVA) and to correlation analysis. The permutation tests can be used for non-normally distributed data and are generally more powerful than analogous non-parametric tests (Manly 1991, Adams and Anthony, 1996).
We obtained personality data (Cattell, 1970) of 196 women tested for latent toxoplasmosis during gravidity. The average intelligence of 158 Toxoplasma free women was 8.3 and of 58 Toxoplasma infected ones was 8.9 on a 10-point scale. The results of F-test showed the difference in variances of infected and uninfected subsets (p=0.048). Therefore, the difference in intelligence must be tested with nonparametric tests. The results (p) of Kolmogorov-Smirnov, Wald-Wolfowitz runs test and Mann-Whitney U test were >0.1, 0.023 and 0.076, respectively.
To test the difference in intelligence between toxoplasma infected and toxoplasma free women with PTPT we prepared the text (ASCII) file TOXO.TXT with 196 lines (one line for every subject) containing two numbers, the intelligence and the code of toxoplasmosis status ("1"- for toxoplasma free and "2"- for toxoplasma infected).
Then we run the program typing a command:
>PTPT -n 5000 -x TOXO.TXT <ENTER>
Within a minute (using a computer 486DX2, 66 Mhz) we obtained the results:
Less: 64 1.22%
Equal: 0 0%
Greater: 4936 98.7%
In an agreement with a theory the permutation test was the most powerful from our battery of statistical tests.
EXAMPLE 3
The correlation between the duration of toxoplasmosis and the amount of a personality shift
The same data set as in the example 2 was used for testing correlation between duration of chronic toxoplasmosis and amount of the shift of a personality factor A (Sizothymia x Affectothymia) (Cattell, 1970). It is known that Toxoplasma infected women have significantly higher factor A (higher Affectothymia), i.e., they are more warmhearted, outgoing, easygoing (Flegr et al., 1996). To find out whether the toxoplasmosis induces the increase of the factor A or whether the women with higher factor A have higher probability of Toxoplasma infection it was necessary to test the correlation between duration of the infection and the change (decrease) of the factor A. The duration of the infection can be assessed on the basis of decrease of anti-toxoplasma antibodies titre. To test the correlation between the antibody titre we prepared a text file ANTIBODY.TXT with 58 lines containing the factor A and antibody titre for every 58 toxoplasma infected women. Then we run the program typing:
>ptpt -n 20000 -c ANTIBODY.TXT
Within a minute we obtained the results:
Less: 337 1.69%
Equal: 3 0.015%
Greater: 19660 98.3%
To eliminate the effect of age of the subjects we can compute age residuals using linear regression between factor A and age of the subject using any statistical program and then preparing the text file RESANTI.TXT containing these residuals instead of factors A. The PTPT analysis of this file provided the results:
Less: 245 1.23%
Equal: 0 0%
Greater: 19755 98.8%
After elimination of the confounding variable age, the result (p=0.0123) was highly significant.
CONCLUSIONS
The program PTPT offers a very powerful and efficient tools for testing a concordance between distribution of phenetic traits and evolutionary history of the taxon. It offers also very powerful tests for statistical analysis of non-normally distributed data. However, with large data sets the time-efficiency of permutation techniques is rather low and rapid computers are necessary to run the analyses. The program PTPT is available at http://www.karlin.mff.cuni.cz/~zaboj/ptpt.
Acknowledgments
We thank Dr. Kulda for providing the pathogenicity data and to Mgr. Vaňáčová for RAPD data. This work was supported by the grant 206/95/0638 of the Grant Agency of Czech Republic.
REFERENCES
Adams D.C. & Anthony C.D. 1996: Using randomization techniques to analyze behavioral data. Anim. Behav. 51: 733-738.
Manly B.F.J. 1991: Randomization and Monte Carlo methods in biology. Chapman and Hall, London.
Archie J.V. 1989: A randomization test for phylogenetic information in systematic data. Syst. Zool. 38: 239-252
Cattell, R.B. (1970) Handbook for the sixteen personality factors questionnaire (16PF), Champain, Institute for Personality and Ability Testing.
Flegr J., Zitkova S., Kodym P. & Frynta D. 1996: Induction of changes in human behaviour by the parasitic protozoan Toxoplasma gondii. Parasitology 113: 49-54.
Harvey P.H. & Pagel M.D. 1991: The comparative method in evolutionary biology. Oxford University Press, Oxford.
Kulda, J. (1989) Employment of experimental animals in studies of Trichomonas vaginalis infection. In: Trichomonads Parasitic in Humans, 205-277. Edited by
Honigberg, B.M. New York, Springer-Verlag.
Manly B.F.J. 1991: Randomization and Monte Carlo methods in biology. Chapman and Hall, London.
Moore J., Freehling M. & Gotelli N.J. 1994: Altered behavior in two species of blattid cockroaches infected with Moniliformis moniliformis (Acanthocephala). J. Parasitol. 80: 220-223.
Thorpe R.S. 1996: The use of DNA divergence to help determine the correlates of evolution of morphological characters. Evolution 50: 524-531.