(for the complete list of publication see link)
Transcription
β-catenin functions as a molecular adapter for disordered cBAF interaction
Chan YS, Gao Q, Robinson SA, Wang W, Filandrova R, Weinhold LM, Cabrera ML, Zhang M, Ambati CSR, Lerario AM, Putluri N, Kiseljak-Vassiliades K, Wierman ME, Habra MA, Hammer GD, Veverka V, Cermakova K, Hodges HC
Mol Cell 2025; DOI: 10.1016/j.molcel.2025.06.026


Structural plasticity of the FOXO-DBD:p53-TAD interaction
Kohoutova K., Srb P., Obsilova V., Veverka V., Obsil T.
Nat Commun 2025; DOI: 10.1038/s41467-025-59106-5

Multivalency of nucleosome recognition by LEDGF
Koutná E, Lux V, Kouba T, Škerlová J, Nováček J, Srb P, Hexnerová R, Šváchová H, Kukačka Z, Novák P, Fábry M, Poepsel S, Veverka V.
Nucleic Acids Research 2023; DOI: 10.1093/nar/gkad674
The TFIIS N-terminal domain (TND): a transcription assembly module at the interface of order and disorder
Čermáková K, Veverka V, Hodges HC.
Biochemical Society Transactions 2023; DOI: 10.1042/bst20220342

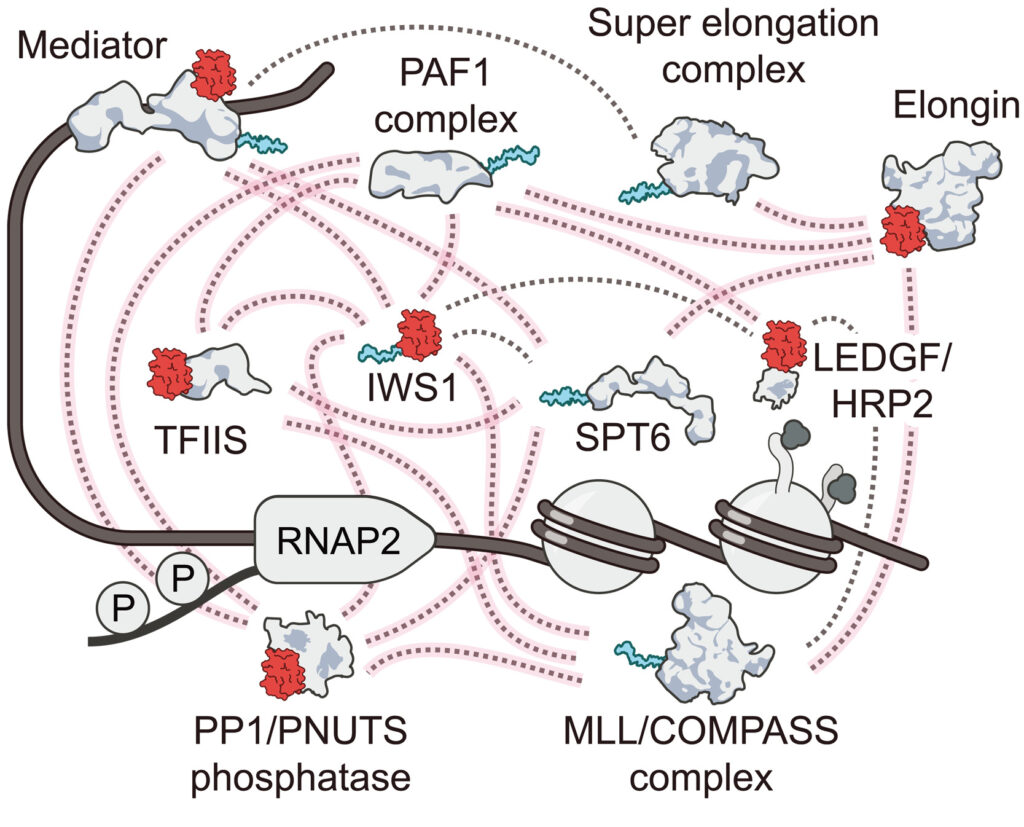
A ubiquitous disordered protein interaction module orchestrates transcription elongation
Čermáková K, Demeulemeester J, Lux V, Nedomova M, Goldman SR, Smith EA, Srb P, Hexnerova R, Fabry M, Madlikova M, Horejsi M, De Rijck J, Debyser Z, Adelman K, Hodges HC, Veverka V.
Science 2021; DOI: 10.1126/science.abe2913
Molecular Mechanism of LEDGF/p75 Dimerization
Lux V, Brouns T, Čermáková K, Srb P, Fábry M, Mádlíková M, Hořejší M, Kukačka Z, Novák P, Kugler M, Brynda J, DeRijck J, Christ F, Debyser Z, Veverka V.
Structure 2020; DOI: 10.1016/j.str.2020.08.012


Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation
Sharma S, Čermáková K, De Rijck J, Demeulemeester J, Fábry M, El Ashkar S, Van Belle S, Lepšík M, Tesina P, Duchoslav V, Novák P, Hubálek M, Srb P, Christ F, Řezáčová P, Hodges HC, Debyser Z, Veverka V.
Proc Natl Acad Sci U S A 2018; DOI: 10.1073/pnas.1803909115
Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers
Hodges HC, Stanton BZ, Cermakova K, Chang CY, Miller EL, Kirkland JG, Ku WL, Veverka V, Zhao K, Crabtree GR.
Nat Struct Mol Biol. 2018; DOI: 10.1038/s41594-017-0007-3
Multiple cellular proteins interact with LEDGF/p75 through a conserved unstructured consensus motif
Tesina P, Čermáková K, Hořejší M, Procházková K, Fábry M, Sharma S, Christ F, Demeulemeester J, Debyser Z, Rijck J, Veverka V, Řezáčová P.
Nat Commun 2015; DOI: 10.1038/ncomms8968
Validation and structural characterization of the LEDGF/p75-MLL interface as a new target for the treatment of MLL-dependent leukemia
Čermáková K, esina P, Demeulemeester J, El Ashkar S, Méreau H, Schwaller J, Rezáčová P, Veverka V, De Rijck J.
Cancer Res 2014; DOI: 10.1158/0008-5472.can-13-3602
Non-canonical DNA
Apollon: a deoxyribozyme that generates a yellow product
Volek M, Kurfürst J, Kožíšek M, Srb P, Veverka V, Curtis EA.
Nucleic Acids Research 2024; DOI: 10.1093/nar/gkae490
Aurora: a fluorescent deoxyribozyme for high-throughput screening
Volek M, Kurfürst J, Drexler M, Svoboda M, Srb P, Veverka V, Curtis EA.
Nucleic Acids Research 2024; DOI: 10.1093/nar/gkae467
Overlapping but distinct: a new model for G-quadruplex biochemical specificity
Volek M, Kolesnikova S, Svehlova K, Srb P, Sgallová R, Streckerová T, Redondo JA, Veverka V, Curtis EA.
Nucleic Acids Research 2021; DOI: 10.1093/nar/gkab037