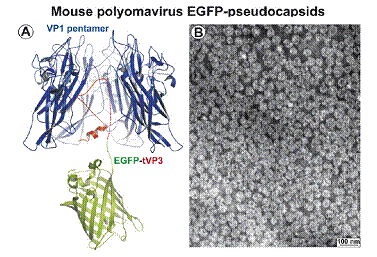
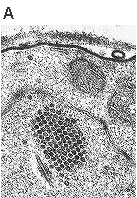
Structure and architecture of the analyzed EGFP capsid-like particles derived from mouse polyomavirus. (A) View-through, showing the macromolecular interaction of VP1 pentamer (depicted in blue) with the C terminus of minor structural protein VP3 (depicted in red) fused with the EGFP protein (depicted in green). (B) Electron microscopy: EGFP-VLPs purified frominsect cell lysates by density gradient were attached to carbon-coated grids, contrasted by negative staining and visualized by electron microscopy. Detailed design, production, properties, and characterization of chimeric EGFP-VLPs were described by Bouřa et al., FEBS Lett 2005.
__________________________________________________________________________________________
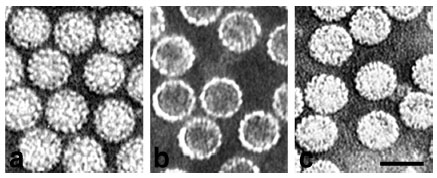
Purified virions (a), empty VP1 pseudocapsids (b), and full VP1 pseudocapsids (c), visualized by electron microscopy (negative staining). Bar, 50 nm. Richterová et al., J Virol 2001.
__________________________________________________________________________________________
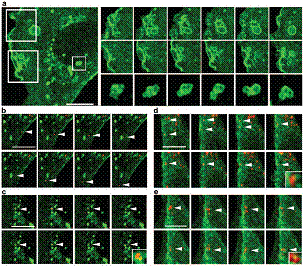
Virus tracking in living NMuMG epithelial cells expressing GFP-fused caveolin-1. (a) Caveolar endocytosis, fission and fusion events, and internal vesicle membrane dynamics are shown not to be affected in a stable cell line. (b) Detail of an infected cell (in a surface confocal section) with a virion bypassing a caveolin-rich domain. (c) In-depth confocal section with a virion(s) captured in a caveolin-rich vesicle at the nuclear periphery. (d and e) Uptake of virions through caveolin membrane domains at the cell surface. Mouse NMuMG epithelial cells expressing GFP-tagged caveolin-1 were infected with Alexa 594-prestained virus (MOI of 10e3 virus particles per cell) and scanned in an open, mediumcontaining chamber with a deltaT of 6 s. Bars, 5 um (a to c) and 2 um (d and e). Liebl et al., J Virol 2006.
__________________________________________________________________________________________
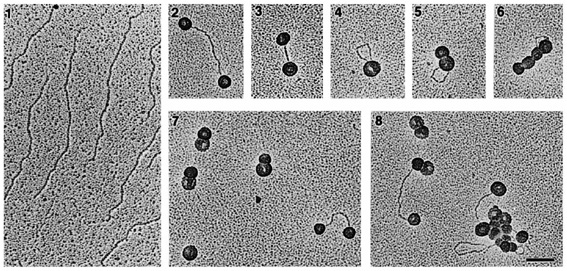
Electron micrographs of interactions of VP1 pseudocapsids with linear DNAs. A: Interactions with: (panel 1) pHC624 DNA (2 kbp) linearised with BamHI, and (panels 2-8) complexed with pseudocapsids. BAC spreading technique, bar = 0.1 um. Štokrová et al., FEBS Lett 1999.
__________________________________________________________________________________________
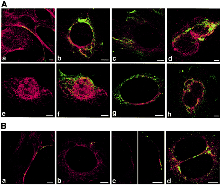
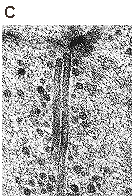
Colocalizations of tubulin with VP1 of pseudocapsids (Aa to Ad) and virions (Ae to Ah and B). Sections of NIH 3T6 fibroblasts (A) and NMuMG (B) cells fixed 20 minutes post infection. (Aa and Ba) or 3 hours post infection (h p.i.) (Ab to Ah and Bb to Bd) were visualized by confocal microscopy. Staining was done with the rabbit anti-polyomavirus virion serum followed by the Alexa Fluor 488-goat anti-rabbit IgG antibody (green) and with the mouse anti-- and anti--tubulin antibody followed by the Cy3-sheep anti-mouse IgG antibody (red). Bars: 5 µm. Richterová et al, J Virol 2001.
__________________________________________________________________________________________
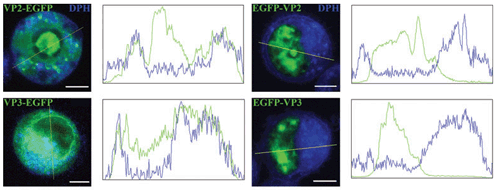
Localization of transiently produced EGFP fused variants of VP2, VP3 minor proteins in living 3T3 cells. Selected confocal microscopy sections of living cells observed 4–5 h post-transfection. Membranes were stained with 1,6-diphenylhexatriene (DPH, blue), EGFP fusion variants of the minor structural proteins are shown in green. Presented profiles of signal intensities were measured across selected lines of shown cell sections. Bars 5 mm. Huerfano et al., FEBS J 2010.
__________________________________________________________________________________________
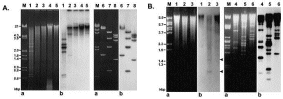
Southern blot analysis of DNA fragments encapsidated by VP1 pseudocapsids in yeast (A) and insect (B) cells. a: Ethidium bromide-stained agarose gel; b: Southern blot. Hybridisation performed with in vitro 32P-labelled DNA isolated from VP1 pseudocapsids produced in yeast (A) and insect (B) cells. A: Restriction fragments of yeast chromosomal DNA (1–4) and pWYGL7L-VP1 plasmid (6–8). Non-digested yeast chromosomal DNA (5); DNAs digested with EcoRI (2, 7), PstI (3), XhoI (4), BamHI, NotI (6) and HindIII (8). B: Restriction fragments of insect cell chromosomal DNA (1–3) and baculovirus DNA (4–6). DNAs digested with EcoRI (1, 4), HindIII (2, 5) and PstI (3, 6). Palková et al., FEBS Letters 2000.
__________________________________________________________________________________________
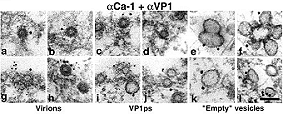
Double staining with the mouse anti-VP1 pseudocapsid polyclonal antibody followed by the 5-nm gold-goat anti-mouse IgG antibody and the rabbit anti-caveolin-1 polyclonal antibody followed by the 10- or 15-nm gold-goat anti-rabbit IgG antibody. Bar, 100 nm (JEOL JEM1200EX). Richterová et al., J Virol 2001.
__________________________________________________________________________________________
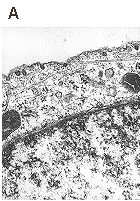
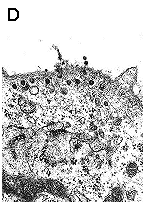
Ultrastructural analysis of the transport pathway of "empty" pseudocapsids (A), "heavy" (B) pseudocapsids and infectious virions (C,D) in mouse fibroblasts 1.5 hour (A,B,C) and 4 hours (D) post infection. Magnification: 60 000x. Liebl D., Diploma thesis 2000
__________________________________________________________________________________________
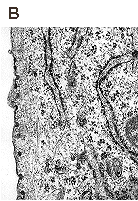
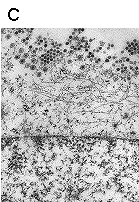
Localization of polyomavirus progeny in the late phases of infection, 36 h p.i. (A), 48 h p.i. (B) and 60 h p.i. (C). Magnification: 60 000x. Liebl D., Diploma thesis 2000
__________________________________________________________________________________________
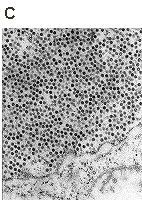
Localization of VP1 pseudocapsids in the nuclei of Saccharomyces cerevisiae cells expressing the VP1 gene. The pseudocapsids are localized either in clusters (A) or dispersed in proximity of structures resembling the mitotic spindle (B,C). Magnification: 84 000x (A,B) and 144 000x (C). Palková et al., FEBS Lett 2000.