Ms1 RNA in bacteria
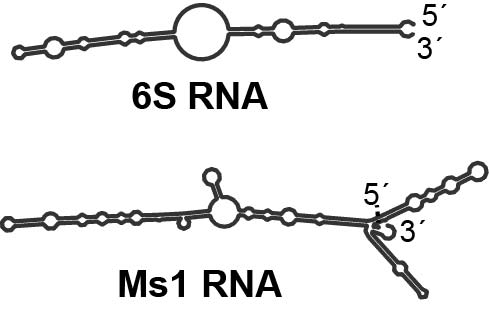
Our research is mainly focused on regulatory RNAs that bind bacterial RNA polymerases (RNAP). In 2013, we discovered that Ms1 RNA, a highly abundant RNA in M. smegmatis stationary phase, binds RNAP core without sigma factors (Hnilicová, 2014). This was the first example of RNA interacting with RNAP in addition to 6S RNA, which binds RNAP holoenzyme with the primary sigma factors. Then, we showed that Ms1 RNA regulates the amount of RNA polymerase in M. smegmatis (Šiková, 2019).
In 2020, we established native RIP-seq to detect any RNA interacting with RNAP in various bacterial species (Vanková Hausnerová, 2024). Collaborating with bioinformaticians, we developed a pipeline to distinguish nascent transcripts associated with transcribing RNAP from RNAs binding to free RNAP. Our findings confirmed that M. smegmatis lacks 6S RNA, but the sigma A and sigma B strongly bind to their respective mRNA transcripts. We started to collaborate with the Czech Army and performed RIP-seq with M. tuberculosis in their BSL3 lab. We showed that the Ms1 homolog, MTS2823, associates with RNAP in the stationary phase. M. smegmatis RIP-seq and our other M. smegmatis NGS data are now publicly available at http://msmegseq.elixir-czech.cz/ web page with an integrated genome browser for better visualization.
RNAP-associated RNAs in actinobacteria
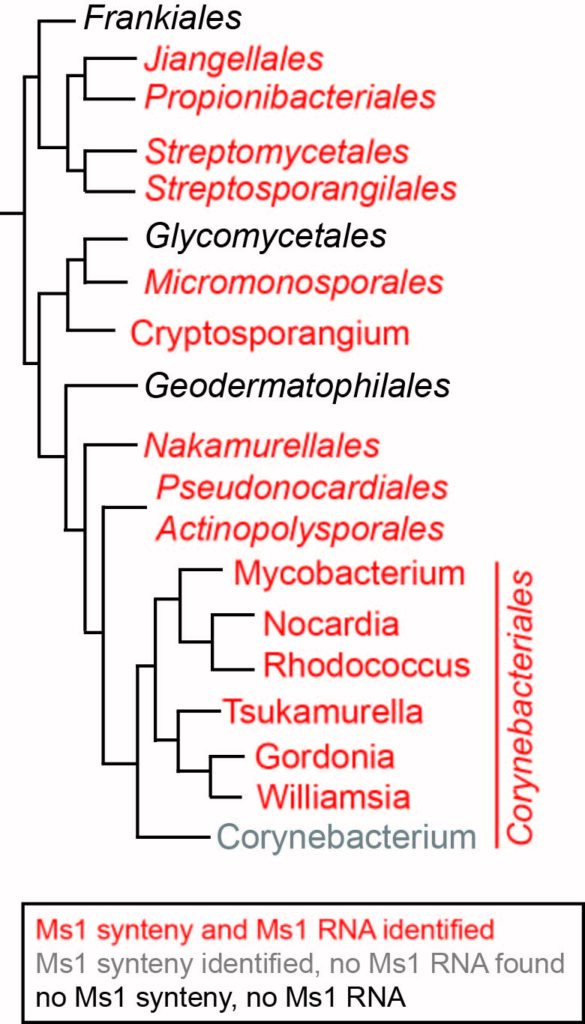
Then, we asked if Ms1 RNA is present in other bacterial species than mycobacteria. We collaborated on this project with Pepa Pánek, a bioinformatician who originally predicted and found Ms1 RNA (Pánek, 2011). By bioinformatic search based on gene synteny, Ms1 length, and secondary structure similarity, we were able to identify Ms1 homologs in many actinobacteria (Vankova Hausnerova, 2022). Later, we confirmed the presence of Ms1 homolog in Streptomyces coelicolor by RIP-seq (Vanková Hausnerová, 2024).
Quite surprisingly, in several species, such as Corynebacterium glutamicum, neither 6S RNA nor Ms1 RNA could be detected by our bioinformatic search (Vankova Hausnerova, 2022). Through RIP-seq, we discovered a novel ~500 nt long RNA binding to both forms of RNAP: RNAP core and RNAP holoenzyme (with the primary sigma factor) in Corynebacterium glutamicum (Vanková Hausnerová, 2024). This RNA has a unique secondary structure and the length distinct from known 6S and Ms1 RNAs, making it undetectable by bioinformatic search. We speculate that other bacteria lacking 6S/Ms1/CoRP RNA might have additional novel types of RNAP-binding RNAs. Identifying these RNAs will enhance our understanding of the diverse RNA structures capable of associating with RNAP.